
| Name | Description | units/level |
| AB | Aboveground biomass | g.plant-1 |
| IRD | Individual fine root density | g.(plant.cell)-1 |
| IRD* | Optimal IRD based on the ESPR | g.(plant.cell)-1 |
| RcB | Coarse root biomass | g.plant-1 |
| RDt | Total fine root density | g.cell-1 |
| RfB | Fine root biomass | g.plant-1 |
| SGr | Net biomass available for somatic growth | g.(day.plant)-1 |
The dynamic allocation module is the core of the model: it will allow plant fine roots to grow according to the ESPR but in a dynamic frame, and then will adjust the other plant compartments of plant anatomy, namely coarse roots and shoot. Note that the allocation into reproduction will be determined by the energy budget module which will run previously and will feed a somatic growth biomass value for each plant x (SGRx) to this module.
Plants that have a negative value of SGrX fed from the energy budget module, meaning that their GPP is superior to their maintenance costs and therefore they must reduce their size, will be considered stressed plants, and will not enter the growth loop. Plant stress history will be tracked so that plants stressed for a number of time steps in a row will die. The rest of plants will enter the plant growth loop (PGL) of this module (Fig 3), where their somatic growth will be simulated (see next paragraph). After all plants leave the PGL because either (i) they have no more somatic growth biomass available (SGR < 0g) or (ii) all the plants remaining in the PGL have been marked as non-growing because they have reached or surpassed their IRD* in all cells (IRDMAX < 0g), the loop for this time step will end. Some plants may the PGL end with an overstock of fine roots because conditions have changed during the loop decreasing their IRD. Therefore, after the loop ends, and IRD decay will be applied: The root surplus will be diminished using a root density dependent decay rate. Finally, RDt will be adjusted accordingly, and the other plant components biomass will also be reduced based on a defined allometry. Any biomass loss during the PGL or further adjustments will be removed from the system. The PGL will be the central component of this module, as it will determine plants’ growth.
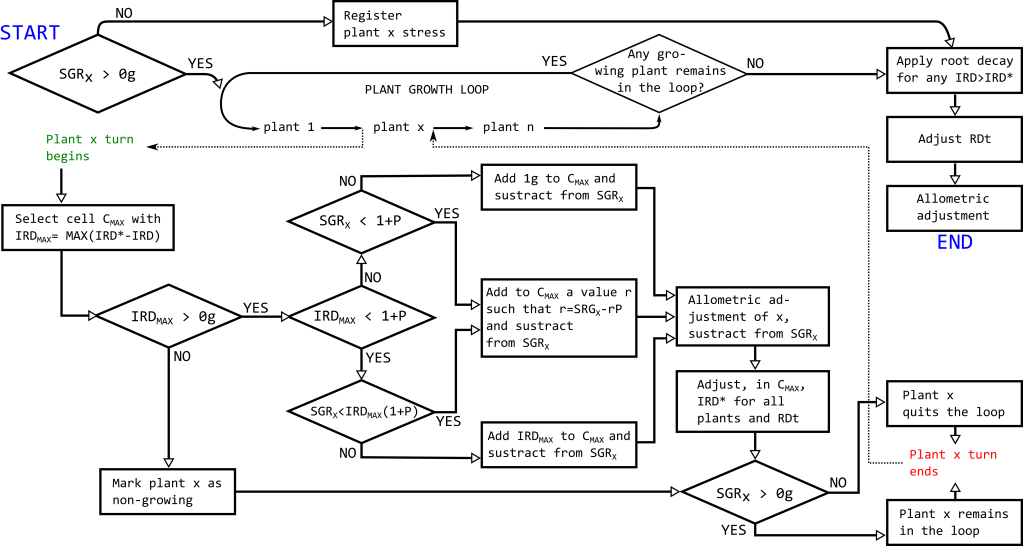
During the PGL each plant will be allowed to grow a limited biomass of fine roots per loop turn (set to 1 gram as an example in Fig 1 and hereafter, but can be adjusted also depending on the time scale decided for each time step) in one grid cell. In every plant turn, first, the cell in which the plant will grow roots, CMAX, will be selected as the grid cell where the difference between current fine root biomass and optimal biomass is maximal (this difference is noted as IRDMAX) among the already occupied cells, or juxtaposed cells hence expanding its root range. During the plant turn, only changes in the cell CMAX will be performed. Second, the amount of biomass allocated into roots in CMAX is determined. If IRDMAX is negative, this means that the plant has reached or surpassed its optimal root growth: Then the plant will be marked as non-growing for this loop run, but the plant will remain in the loop because it may find eventually beneficial to grow some roots as a response to other plant roots’ proliferation. If IRDMAX is positive, this means that there is at least one grid cell where the plants benefits from root proliferation according to the ESPR. In this case, an amount r of biomass will be added to the value of IRD in CMAX, accounting for the proportional amount of biomass P necessary to fulfill the allometric growth of coarse roots and shoot. If SGrX constrains growth (is lower than what the plant would grow according to IRDMAX), all the SGrX will be allocated to growth and the plant will eventually quit the loop (SGRX will become 0). If growth is not constrained by SGrX, the plant will grow its IRD in CMAX a maximum amount of 1g (for example) of biomass, but if IRDMAX is lower than this value then it will grow IRDMAX. After fine root proliferation, and before ending the plant’s turn in the loop, r.P biomass will be allocated to coarse roots and to shoot and subtracted from SGrX, and IRD for each plant and RDt will be updated in CMAX. In each loop run all plants will go thru this process. By the end on a loop run the plants state will be evaluated, and if all plants in the loop are marked as non-growing the loop ends, otherwise it will run again.